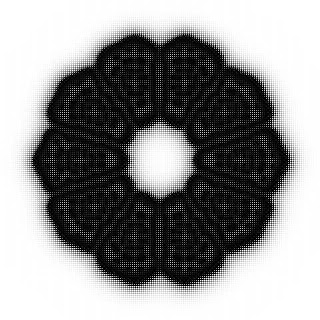
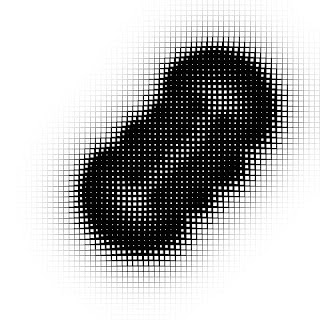
Creative coding on Turing pattern
It's an article that describes the creative coding with the 'p5.js' and the 'Processing' to create the unique and mysterious Turing pattern.
It contains the points to create interesting patterns, the simple example code, and the application example code. You can surely make your original work with these.
What is the Turing pattern?
Simply speaking, the Turing pattern is the pattern that looks like the surface of tropical fish. And the Turing pattern is generated with the reaction-diffusion equations.
English mathematician Alan Turing researched the reaction-diffusion theory.
Wikipedia : Turing pattern
It needs to do many calculations to draw the Turing pattern. And that is where the computer is at its strongest.
Basic example code of 'p5.js'.
Here is the basic code of 'p5.js' to draw the Turing pattern.
/*
* Reaction-Diffusion system by the Gray-Scott Model.
* basic example.
*
* @author @deconbatch
* @version 0.1
* p5.js 1.1.3
* license CC0
* created 2022.03.26
*/
const w = 480;
const h = w;
const cSiz = 3; // cell size
const pCnt = 500; // calculation count
function setup() {
createCanvas(w, h);
noLoop();
const lab = new Labo(cSiz);
lab.init();
for (let i = 0; i < pCnt; i++) {
lab.proceed();
}
lab.observe();
}
/*
* Labo : reaction-diffusion system.
*/
class Labo {
cellSize;
matrixW;
matrixH;
diffU;
diffV;
cells;
constructor(_cSiz) {
this.cellSize = _cSiz;
this.matrixW = floor(width / this.cellSize);
this.matrixH = floor(height / this.cellSize);
this.diffU = 0.9;
this.diffV = 0.1;
this.cells = new Array();
}
/*
* init : initialize reaction-diffusion system.
*/
init() {
for (let x = 0; x < this.matrixW; x++) {
this.cells[x] = [];
for (let y = 0; y < this.matrixH; y++) {
this.cells[x][y] = new Cell(
map(x, 0.0, this.matrixW, 0.03, 0.12), // feed
map(y, 0.0, this.matrixH, 0.045, 0.055), // kill
1, // u
(random(1) < 0.1) ? 1 : 0 // v
);
}
}
}
/*
* proceed : proceed reaction-diffusion calculation.
*/
proceed() {
// calculate Laplacian
const nD = Array(); // neighbors on diagonal
const nH = Array(); // neighbors on vertical and horizontal
for (let x = 0; x < this.matrixW; x++) {
for (let y = 0; y < this.matrixH; y++) {
// set neighbors
nD[0] = this.cells[max(x-1,0)][max(y-1,0)];
nD[1] = this.cells[max(x-1,0)][min(y+1,this.matrixH-1)];
nD[2] = this.cells[min(x+1,this.matrixW-1)][max(y-1,0)];
nD[3] = this.cells[min(x+1,this.matrixW-1)][min(y+1,this.matrixH-1)];
nH[0] = this.cells[max(x-1,0)][y];
nH[1] = this.cells[x][max(y-1,0)];
nH[2] = this.cells[x][min(y+1,this.matrixH-1)];
nH[3] = this.cells[min(x+1,this.matrixW-1)][y];
// Laplacian
let c = this.cells[x][y];
let sum = 0.0;
for (let i = 0; i < 4; i++) {
sum += nD[i].valU * 0.05 + nH[i].valU * 0.2;
}
sum -= c.valU;
c.lapU = sum;
sum = 0.0;
for (let i = 0; i < 4; i++) {
sum += nD[i].valV * 0.05 + nH[i].valV * 0.2;
}
sum -= c.valV;
c.lapV = sum;
}
}
// reaction-diffusion
for (let x = 0; x < this.matrixW; x++) {
for (let y = 0; y < this.matrixH; y++) {
let c = this.cells[x][y];
let reaction = c.valU * c.valV * c.valV;
let inflow = c.feed * (1.0 - c.valU);
let outflow = (c.feed + c.kill) * c.valV;
c.valU = c.valU + this.diffU * c.lapU - reaction + inflow;
c.valV = c.valV + this.diffV * c.lapV + reaction - outflow;
c.standardization();
}
}
}
/*
* observe : display the result.
*/
observe() {
background(0);
fill(255);
noStroke();
for (let x = 0; x < this.matrixW; x++) {
for (let y = 0; y < this.matrixH; y++) {
let cx = x * this.cellSize;
let cy = y * this.cellSize;
let cs = this.cells[x][y].valU * this.cellSize;
rect(cx, cy, cs, cs);
}
}
}
}
/*
* Cell : holds cell informations.
*/
class Cell {
feed;
kill;
valU;
valV;
lapU;
lapV;
constructor(_f, _k, _u, _v) {
this.feed = _f;
this.kill = _k;
this.valU = _u;
this.valV = _v;
this.lapU = 0;
this.lapV = 0;
}
standardization() {
this.valU = constrain(this.valU, 0, 1);
this.valV = constrain(this.valV, 0, 1);
}
}
Notice : When you try this on OpenProcessing, care about 'loop protection'.
This code contains the reaction-diffusion system called the 'Gray-Scott' model. And the pattern changes with the parameters on the reaction-diffusion equation on the XY axis.
Brief description of the code.
It calculates the reaction-diffusion equations in 'class Labo'. The calculation scope is 'matrixW x matrixH' area. 'calss Cell' symbolizes the square on the matrix. And 'cell[matrixW][matrixH]' stands for the whole squares in the calculation scope.
'class Labo init()' initializes the whole squares in the calculation scope. 'class Labo proceed()' calculates the reaction-diffusion equations once. The caller controls how many times to call the calculation.
The calculation amount of this code is quite much. So it takes a long time to get a result. The calculation time is proportional to 'pCnt', and it will be four times if 'cSize' was halved.
The parameters below took about one minute in my environment.
const cSiz = 3; // cell size
const pCnt = 500; // calculation count
The two-dimensional array 'this.cells[x][y]' hold the calculation results. And 'class Labo observe()' draws with it.
Though example code draws monochrome image, you can develop any way you like.
Creative coding points to create interesting patterns.
Tweak the parameter values.
The looks of the pattern depend on the parameter values of the reaction-diffusion equations.
The example code uses four parameters in the two parts below. It sets 'diffU' and 'diffV' to constant values, and sets 'feed' and 'kill' the values related to the XY axis location.
// diffU, diffV
this.diffU = 0.9;
this.diffV = 0.1;
// feed, kill
this.cells[x][y] = new Cell(
map(x, 0.0, this.matrixW, 0.03, 0.12), // feed
map(y, 0.0, this.matrixH, 0.045, 0.055), // kill
For example, setting 'feed' and 'kill' to the 'A' and 'B' related values show characteristic patterns like this.
The example code will create the patterns when the values of 'feed' and 'kill' are in a range like these.
feed : 0.03 - 0.12
kill : 0.045 - 0.055
You can change the values of 'diffU' and 'diffV'. But it will be hard to get some patterns.
Plot the seeds.
The example code set 'V' values in the calculation scope to '1' randomly.
this.cells[x][y] = new Cell(
(random(1) < 0.1) ? 1 : 0 // v
You can change the calculation result with the way to set the 'V' values.
You can set the 'V' values in 'class Labo init()'. At first, set the whole 'V' values to '0' and set the part of 'V' values to '1' you want to set.
// Example to set circular.
for (let t = 0; t < TWO_PI; t += PI * 0.2) {
let x = floor(this.matrixW * (0.5 + 0.25 * cos(t)));
let y = floor(this.matrixW * (0.5 + 0.25 * sin(t)));
this.cells[x][y].valV = 1;
}
Calculation count and the square size.
The calculation count of reaction-diffusion equations and the square size in the calculation scope affect the look of the pattern.
const cSiz = 3; // cell size
const pCnt = 1000; // calculation count
Creative coding application example of the Turing pattern.
These are the creative coding application example based on the points to create the pattern shown above. I'll share these with the CC0 license.
The 'p5.js' application example code.
Change 'class Labo init()' below.
/*
* init : initialize reaction-diffusion system.
*/
init() {
const hW = floor(this.matrixW * 0.5);
const hH = floor(this.matrixH * 0.5);
for (let x = 0; x < this.matrixW; x++) {
this.cells[x] = [];
for (let y = 0; y < this.matrixH; y++) {
let d = dist(x, y, hW, hH);
let f = map(sin(TWO_PI * d * 3 / hW), -1, 1, 0.12, 0.03);
this.cells[x][y] = new Cell(
f, // feed
0.045, // kill
1, // u
0 // v
);
}
}
for (let t = 0; t < TWO_PI; t += PI * 0.2) {
for (let r = 0.1; r < 0.4; r += 0.1) {
let x = floor(this.matrixW * (0.5 + r * cos(t)));
let y = floor(this.matrixW * (0.5 + r * sin(t)));
this.cells[x][y].valV = 1;
}
}
}
The 'Processing' application example code.
It's the same application example written in the 'Processing'.
<
/**
* Reaction-Diffusion system by the Gray-Scott Model.
* application example.
*
* @author @deconbatch
* @version 0.1
* Processing 3.5.3
* license CC0
* created 2022.03.26
*/
void setup() {
size(480, 480);
noLoop();
int cSiz = 2; // cell size
int pCnt = 1000; // calculation count
Labo lab = new Labo(cSiz);
lab.init();
for (int i = 0; i < pCnt; i++) {
lab.proceed();
}
lab.observe();
}
/*
* Labo : reaction-diffusion system.
*/
public class Labo {
int cellSize;
int matrixW;
int matrixH;
float diffU;
float diffV;
Cell[][] cells;
Labo(int _cSiz) {
cellSize = _cSiz;
matrixW = floor(width / cellSize);
matrixH = floor(height / cellSize);
diffU = 0.9;
diffV = 0.1;
cells = new Cell[matrixW][matrixH];
}
/*
* init : initialize reaction-diffusion system.
*/
void init() {
float hW = matrixW * 0.5;
float hH = matrixH * 0.5;
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
float d = dist(x, y, hW, hH);
float f = map(sin(TWO_PI * d * 3.0 / hW), -1.0, 1.0, 0.03, 0.12);
cells[x][y] = new Cell(
f, // feed
0.045, // kill
1.0, // u
0.0 // v
);
}
}
for (float t = 0.0; t < TWO_PI; t += PI * 0.2) {
for (float r = 0.1; r < 0.4; r += 0.1) {
int x = floor(matrixW * (0.5 + r * cos(t)));
int y = floor(matrixW * (0.5 + r * sin(t)));
cells[x][y].setV(1.0);
}
}
}
/*
* proceed : proceed reaction-diffusion calculation.
*/
void proceed() {
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
// neighbors on diagonal
Cell[] nD = new Cell[4];
nD[0] = cells[max(x-1,0)][max(y-1,0)];
nD[1] = cells[max(x-1,0)][min(y+1,matrixH-1)];
nD[2] = cells[min(x+1,matrixW-1)][max(y-1,0)];
nD[3] = cells[min(x+1,matrixW-1)][min(y+1,matrixH-1)];
// neighbors on vertical and horizontal
Cell[] nH = new Cell[4];
nH[0] = cells[max(x-1,0)][y];
nH[1] = cells[x][max(y-1,0)];
nH[2] = cells[x][min(y+1,matrixH-1)];
nH[3] = cells[min(x+1,matrixW-1)][y];
// lapU
Cell c = cells[x][y];
float sum = 0.0;
for (int i = 0; i < 4; i++) {
sum += nD[i].getU() * 0.05 + nH[i].getU() * 0.2;
}
sum -= c.getU();
c.setLapU(sum);
// lapV
sum = 0.0;
for (int i = 0; i < 4; i++) {
sum += nD[i].getV() * 0.05 + nH[i].getV() * 0.2;;
}
sum -= c.getV();
c.setLapV(sum);
}
}
// reaction-diffusion
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
Cell c = cells[x][y];
float reaction = c.getU() * c.getV() * c.getV();
float inflow = c.getFeed() * (1.0 - c.getU());
float outflow = (c.getFeed() + c.getKill()) * c.getV();
c.setU(c.getU() + diffU * c.getLapU() - reaction + inflow);
c.setV(c.getV() + diffV * c.getLapV() + reaction - outflow);
c.standardization();
}
}
}
/*
* observe : display the result.
*/
void observe() {
background(0);
fill(255);
noStroke();
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
int cx = x * cellSize;
int cy = y * cellSize;
float cs = cells[x][y].getU() * cellSize;
rect(cx, cy, cs, cs);
}
}
}
}
/**
* Cell : hold the informations of the cell.
*/
public class Cell {
private float feed;
private float kill;
private float valU;
private float valV;
private float lapU;
private float lapV;
Cell(float _f, float _k, float _u, float _v) {
feed = _f;
kill = _k;
valU = _u;
valV = _v;
lapU = 0.0;
lapV = 0.0;
}
public void setLapU(float _l) {
lapU = _l;
}
public void setLapV(float _l) {
lapV = _l;
}
public void setU(float _u) {
valU = _u;
}
public void setV(float _v) {
valV = _v;
}
public float getFeed() {
return feed;
}
public float getKill() {
return kill;
}
public float getU() {
return valU;
}
public float getV() {
return valV;
}
public float getLapU() {
return lapU;
}
public float getLapV() {
return lapV;
}
public void standardization() {
valU = constrain(valU, 0.0, 1.0);
valV = constrain(valV, 0.0, 1.0);
}
}